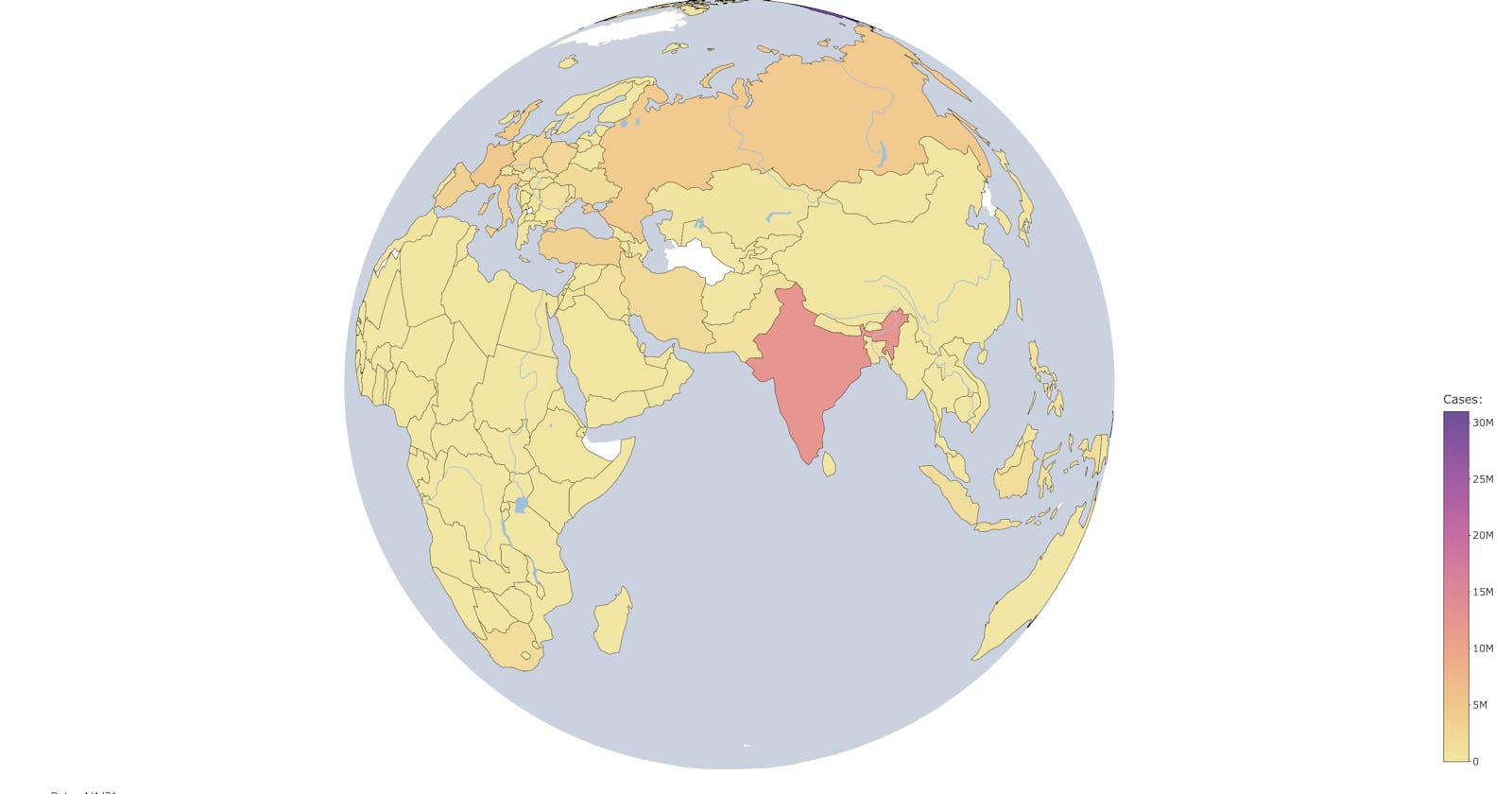
Building interactive visualizations in R is quickly becoming a popular method of visualizing large and complex datasets. In recent months, I have been fascinated by trends in COVID-19 data. Using this as inspiration, this post will be describing how to build an interactive globe in R for visualizing time-series data. In this tutorial, the globe will be both interactive (meaning that it can be moved and panned around on screen, with countries being able to be hovered over for more information), and a slider will be present at the bottom of the screen to move the globe “through time” in the dataset.
To create this visualization, you'll need the following packages. These can easily be installed with the install.packages() command.
library(plotly)
library(countrycode)
library(dplyr)
Next, you’ll need to load the data into R. For this visualization I used COVID-19 global case count data from John Hopkins University. In this dataset, each row is data from an individual country, and each column is the cumulative case count for that particular day.
global_case_data <- read.csv('https://github.com/CSSEGISandData/COVID-19/raw/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv')
In this dataset specifically, the United States was denoted as ‘US’. Given that the rest of the countries in this dataset were denoted by their full name, I decided to rename this data point for consistency. This step is entirely optional.
global_case_data$Country.Region[250] <- "United States"
Another quirk of this dataset is that it has two country columns. The first denotes the state or province, the second the country itself. For simplicity, I combined all the rows that were split over different states/provinces, using the code below.
First, I took the second column of the data frame, the country column, as a list, and iterated through it. I then subsetted the data frame to contain just the rows pertaining to that country. If the number of rows in that subsetted data frame was equal to one, then that row was appended to a new data frame (gc_data). However, if the number of rows in the subsetted data frame was greater than one, I then summed the columns of each row to acquire a total day-by-day case count for all the states/provinces in that country.
gc_data <- data.frame()
for (country in unique(global_case_data$Country.Region)) {
subset <- global_case_data %>% filter(Country.Region == country)
temp <- subset[1,]
if (nrow(subset) > 1) {
for (i in 2:nrow(subset)) {
temp[,5:ncol(subset)] <- temp[,5:ncol(subset)] + subset[i,5:ncol(subset)]
}
}
gc_data <- rbind(gc_data, temp)
}
Once the consolidation of the data was done, an extra column was appended to the case count data frame that contained the country code of each country in the dataset. This becomes relevant later on, as the country codes are needed to map the data onto the correct country.
gc_data$Codes <- countrycode(gc_data$Country.Region, origin = 'country.name',
destination = 'iso3c')
gc_data <- rbind(gc_data, rep(FALSE, ncol(gc_data)))
Now, specify the time intervals that you wish to be present in your slider. I picked the first of every month, starting from March 1st 2020, all the way to April 1st 2021.
dates = c('X3.1.20', 'X4.1.20', 'X5.1.20', 'X6.1.20', 'X7.1.20',
'X8.1.20', 'X9.1.20', 'X10.1.20', 'X11.1.20', 'X12.1.20',
'X1.1.21', 'X2.1.21', 'X3.1.21','X4.1.21')
To make the slider look nicer later on, I created labels for each of the dates.
# Create print-ready labels for figure
date_str = c('3/1/20', '4/1/20', '5/1/20', '6/1/20', '7/1/20',
'8/1/20', '9/1/20', '10/1/20', '11/1/20', '12/1/20',
'1/1/21', '2/1/21', '3/1/21', '4/1/21')
Last (in terms of pre-processing the data), you want to ensure that the first date shown on the figure is visible by default (this is explained in more detail later).
gc_data[nrow(gc_data), dates[1]] = TRUE
In order to create a readable choropleth map, Plotly uses a series of settings that are defined as a list. These settings can of course be tweaked to however you want to create your map, however the following settings were used for the map. The only one that shouldn’t be changed (unless you are not aiming for a globe map view) is the projection = list(type = 'orthographic’) setting. This is what creates a globe instead of another map view. Click here for a list of other map views in Plotly.
border_settings <- list(color = toRGB("#000000"), width = 0.5)
globe_settings <- list(
showframe = FALSE,
showcoastlines = FALSE,
projection = list(type = 'orthographic'),
resolution = '100',
showcountries = TRUE,
countrycolor = '#d1d1d1',
showocean = TRUE,
oceancolor = '#c9d2e0',
showlakes = TRUE,
lakecolor = '#99c0db',
showrivers = TRUE,
rivercolor = '#99c0db')
After defining the settings for the map, I create the initial, generic map object. Right now, this plot object (named fig has no settings or context for the data).
fig <- plot_geo(gc_data)
The next block of code is the most complex part of this tutorial. As mentioned above, not every single day in the dataset will be visualized. The next step builds the map overlays for each time slice that I am visualizing. First I create a list to store visibility information for each time slice. The reason for this becomes more apparent later. Then I iterate through the previously defined list of dates via a loop. Every iteration of the loop does a few things. First its important to understand how Plotly handles adding data to a map. It does so via a “trace” which acts as an overlay of data onto countries. Each iteration of this loop, adds a trace by pulling the proper data from the proper date (via the current loop index). It also sets the visibility of that trace to whatever was placed on the bottom of the rows previously (not visible, except for the initial trace). It applies a uniform color scheme to the trace, I used “Sunset” and it turns the Country Region column of the data into the map labels. To map a country’s data to the proper portion of the map, the country codes found earlier is used for the locations parameter. Finally the border settings defined earlier are applied. Additionally, using the %>% operator, a title, map legend, and the map settings that I created earlier.
This loop also manages the visibility of the layers. As mentioned, I created a steps list that will store the visibility of the traces as I slide through the visualization. This section creates a series of nested lists, that set all visibilities but the specific trace to be visible. This allows for turning off traces as a new time is selected by the slider.
steps <- list()
for (i in 1:length(dates)) {
fig <- fig %>% add_trace(z = ~gc_data[, dates[i]],
color = ~gc_data[, dates[i]],
visible = as.logical(gc_data[nrow(gc_data), dates[i]]),
colors = hcl.colors(10, "Sunset", rev=TRUE),
text = ~Country.Region,
locations = ~Codes,
marker = list(line = border_settings)) %>%
layout(title = 'COVID-19 Cases Over Time', geo = globe_settings) %>%
colorbar(title = 'Cases:', limits = c(0, 31000000), len=.5, which=i, y=.5)
step <- list(args = list('visible', rep(FALSE, length(dates))),
method = 'restyle', label=date_str[i])
step$args[[2]][i] = TRUE
steps[[i]] = step
}
Finally, I need to add the slider, and tie it to the figure. I do that with the following code. I can label the slider with the “current value” and prefix functionality, and steps can be passed in to modify visibility.
fig <- fig %>%
layout(sliders = list(list(active = 0,
currentvalue = list(prefix = "Date: "),
steps = steps)))
From this point, the visualization is done. It can be viewed by simply calling fig in the code or R console. If you intend to save the visualization, save it as an HTML document, as this will keep the interactive component.

Complete Code
Running this will generate an interactive globe as intended by the tutorial. There are some memory management function calls that improve memory usage of this script that was not included in the tutorial. They are not strictly necessary, however, they improve the performance of the script on low-powered machines.
library(plotly)
library(countrycode)
library(dplyr)
# Global Case Count
global_case_data <- read.csv('https://github.com/CSSEGISandData/COVID-19/raw/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv')
# Rename 'US' to 'United States' for consistency
global_case_data$Country.Region[250] <- "United States"
# Consolidate data - combining rows that are split over different regions of a country
gc_data <- data.frame()
for (country in unique(global_case_data$Country.Region)) {
subset <- global_case_data %>% filter(Country.Region == country)
temp <- subset[1,]
if (nrow(subset) > 1) {
for (i in 2:nrow(subset)) {
temp[,5:ncol(subset)] <- temp[,5:ncol(subset)] + subset[i,5:ncol(subset)]
}
}
gc_data <- rbind(gc_data, temp)
}
# Clean up memory after preprocessing
rm(temp, subset, country, i)
gc()
# Append a column for country codes
gc_data$Codes <- countrycode(gc_data$Country.Region, origin = 'country.name',
destination = 'iso3c')
gc_data <- rbind(gc_data, rep(FALSE, ncol(gc_data)))
# Identify column headers to query for slider
dates = c('X3.1.20', 'X4.1.20', 'X5.1.20', 'X6.1.20', 'X7.1.20',
'X8.1.20', 'X9.1.20', 'X10.1.20', 'X11.1.20', 'X12.1.20',
'X1.1.21', 'X2.1.21', 'X3.1.21','X4.1.21')
# Create print-ready labels for figure
date_str = c('3/1/20', '4/1/20', '5/1/20', '6/1/20', '7/1/20',
'8/1/20', '9/1/20', '10/1/20', '11/1/20', '12/1/20',
'1/1/21', '2/1/21', '3/1/21', '4/1/21')
# Ensure that first frame is visible
gc_data[nrow(gc_data), dates[1]] = TRUE
# create steps and plot all traces
# Set country boundaries as light grey
border_settings <- list(color = toRGB("#000000"), width = 0.5)
# Specify map projection and options
globe_settings <- list(
showframe = FALSE,
showcoastlines = FALSE,
projection = list(type = 'orthographic'),
resolution = '100',
showcountries = TRUE,
countrycolor = '#d1d1d1',
showocean = TRUE,
oceancolor = '#c9d2e0',
showlakes = TRUE,
lakecolor = '#99c0db',
showrivers = TRUE,
rivercolor = '#99c0db')
# Create initial plot object
fig <- plot_geo(gc_data)
# Iterate across time slices and add individual globe figures
# to the figure so that a slider can slide across the date range
# also creates steps so that when the slider is used, visibility for
# other globes are disabled
steps <- list()
for (i in 1:length(dates)) {
# Specifically, it maps global covid data to the z function (a layer) on the map
fig <- fig %>% add_trace(z = ~gc_data[, dates[i]],
color = ~gc_data[, dates[i]],
visible = as.logical(gc_data[nrow(gc_data), dates[i]]),
colors = hcl.colors(10, "Sunset", rev=TRUE),
text = ~Country.Region,
locations = ~Codes,
marker = list(line = border_settings)) %>%
layout(title = 'COVID-19 Cases Over Time', geo = globe_settings) %>%
colorbar(title = 'Cases:', limits = c(0, 31000000), len=.5, which=i, y=.5)
# This step is what determines layer visibility
# as the slider moves through the dates
step <- list(args = list('visible', rep(FALSE, length(dates))),
method = 'restyle', label=date_str[i])
step$args[[2]][i] = TRUE
steps[[i]] = step
}
# Add slider control to plot
fig <- fig %>%
layout(sliders = list(list(active = 0,
currentvalue = list(prefix = "Date: "),
steps = steps)))
# Display Figure
fig
# Remove unwanted variables to ensure memory cleanup
rm(border_settings, fig, gc_data, global_case_data,
date_str, dates, i, globe_settings, step, steps)
gc()